Molecular detection of Trypanosoma evansi using RoTat 1.2 (VSG) genes in cattles from Borno and Gombe States, Nigeria
DOI:
https://doi.org/10.51607/22331360.2024.73.3.181Keywords:
Cattle, Nigeria, RoTat 1.2 Gene, Trypanosoma evansiAbstract
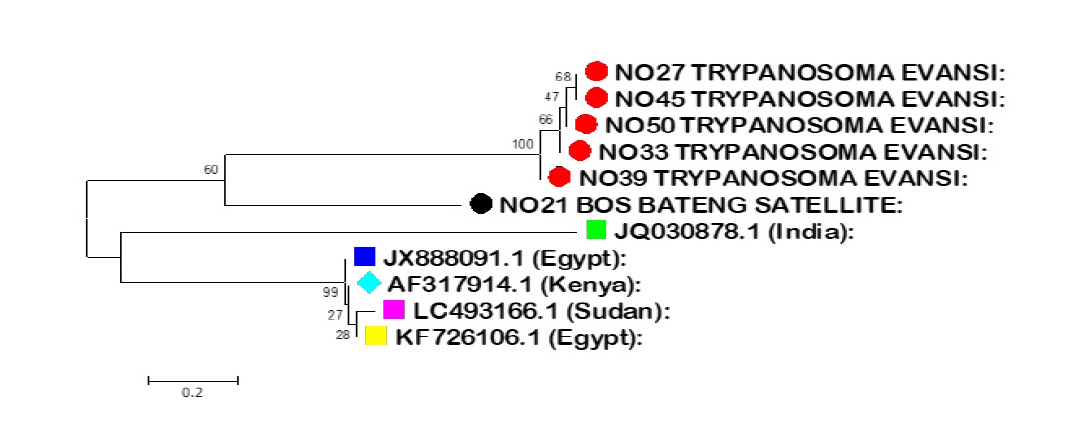
Trypanosoma evansi (T. evansi) causes Surra, an important haemoprotozoan disease of economic importance that affect wide range of host. The research was designed to detection of T. evansi using molecular method in cattle from Gombe and Borno States, Nigeria. Blood samples from apparently four hundred and fifty-five (455) cattle were collected from the month of May to October 2018 in different Local Government areas. Microscopy detection revealed an overall prevalence rate of 11.8% in Gombe State and 9.0% in Borno State during the study period. Out of the 455 blood samples collected, fifty (50) samples, with 25 each from Borno and Gombe states, were subjected to molecular analysis for identification of Trypanosoma evansi. Twenty-one (21) positive sample of T. evansi was detected in 50 cattle, tested using PCR targeting the RoTat 1.2 variable surface glycoproteins (VSG) gene. PCR-positive samples were further phylogenetically analysed. Sequence analysis of the Rotat 1.2 VSG gene showed high variability between isolates from this study and the isolates earlier deposited in the Genbank. Microscopic examination exhibited very low sensitivity, whereas PCR using RoTat 1.2 VSG gene showed more sensitivity in the diagnosis of T. evansi from cattle in Gombe and Borno States. Conclusively, the phylogenies results obtained from the Neighbor joining technique suggest that the isolate of T. evansi in this study are not related to the isolates from Egypt, Kenya, Sudan and India.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Amina Mohammed Bello, Hassan Abdulselam, Jamila Dauda, Salamatu Mohammed Tukur, Hauwa Ibrahim Abdulrahman, Hussaina Maidugu Bata Maidala, Lawan Adamu, Modu Muhammad Bukar, Albert Wulari Mbaya, Godwin Onyemaechi Egwu

This work is licensed under a Creative Commons Attribution 4.0 International License.







